``
Hello and welcome to week 7 of ESPM 112L-
Metagenomic Data Analysis Lab!
- What lists are for
- Using Custom ggKbase Lists
- Optional: Download sequences for analysis
- Today’s turn-in assignment:
This week we’re going to be investigating the metabolic capacities of the organisms in your samples. This is a great chance to get to know your bacteria (and viruses) a little better!
The main way we’re going to be looking at this information this week is by using ‘genome summaries’, which are a tool included in the ggkbase platform.
Genome summaries are an important part of the ggkbase platform that allow users to interrogate the metabolic potential of the bins they’ve created, and see what remains in the unbinned fraction. To make a genome summary, first click on Genome Summary at the top of the page, then click Create Genome Summary.
At this point you are able to restrict genome summaries to certain projects. For the purposes of this assignment, select all projects you have access to and click “Go to Genome Summary”.
You are now at your new, blank, genome summary. Now, click Select organisms. You can now choose the organisms that you would like to include in this figure. Again, for the purposes of this assignment, click Select All Organisms in the top left of the popup window, and then Apply.
First, we’ve recently added a new system that uses HMMs to detect genes, but it’s not been run on this dataset, so we’ll need to stop it from displaying. Click “Select HMMs” (right-most green button), then at the top left click “Deselect all HMMs”, then “Apply”. Now we can start from scratch.
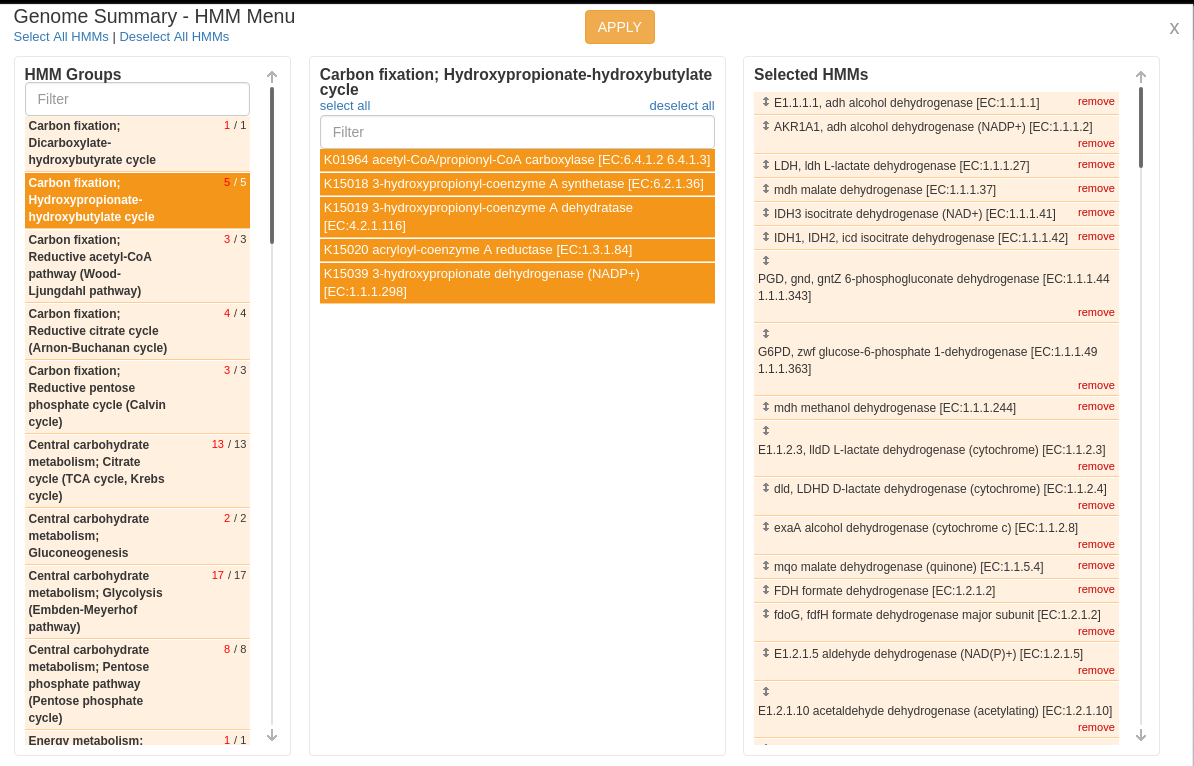
Now click “Select lists”. A screen now pops up showing you all of the “universal lists” which are populated when the data are imported. They’re grouped first by category, and then by smaller groups within each category. Pick some lists which interest you- some especially interesting ones to look at are the electron transport chain (complexes I-IV), fermentative metabolism, and important biogeochemical cycles of hydrogen, sulfur, and nitrogen. Some (not all!) of the genes involved in these pathways are shown below:

This is your genome summary! It shows each organism you selected as rows, and the number of proteins in each organism that match the search terms in each list. So in the screen-shot above, you can see that many organisms in your sample have fermentative genes (pink) and lack many electron transport chains (brown), suggesting that these are probably anaerobes. As you might expect, being that they live inside a cow gut!
To save this summary you made, simply give it a name by clicking on “Choose name” (by “Untitled”). Now you can leave the page, and get back to this summary by clicking Genome Summary at the top of ggkbase again.
To investigate the proteins in each category, you can click on the numbers within the boxes. This will show the list of features which hit this list. From this list, you can find out which scaffold they’re on, they’re location, the sequence (for BLAST searches), the annotations, and more!
What lists are for
Genome summaries and lists are intended to make it easier to visualize metabolic capacities across a large number of organisms and to share these visualizations with other researchers. You can make custom lists, as you’ll learn about shortly, and can create genome summaries for various groups of genomes, making this a wonderful tool not only for extracting insights from your data but also to convey these insights to others.
Once you’ve given the genome summary a name and saved it, you can then share the link to other people with access to ggkbase, who can then view your summary. Very useful for group projects!
Using Custom ggKbase Lists
We just used universal lists in our genome summary. Lists work by using key terms. ggKbase searches these terms against all of the annotations in all called proteins in all selected projects. As shown in the List Keywords section, there is some ability to refine you lists to your liking using these keywords.
There is a fantastic help page set up to introduce you to lists on ggkbase, and give you some tips and tricks: <a href=http://ggkbase-help.berkeley.edu/analysis/lists/>http://ggkbase-help.berkeley.edu/analysis/lists/</a>
To make your own list, click on Lists at the top of the page. Next, click Create a new list on the top right of the page. From here you can fill in all of the details to make your own list. You can give it a name, color, and description. The most important part of the list are the terms you select to include and exclude. (This will be covered in the video for today’s lab in more detail- go watch that!)
The three search boxes use boolean logic. The first box produces genes that have annotations matching one or more of the keywords (boolean OR). The second search box requires the genes annotation to match all the keywords (boolean AND). The last search box allows you to enter undesired keywords, excluding genes that have these keywords in their annotations (boolean NOT).

Let’s try, as an example, making a list that just looks at Nitrate reductase, a family of related genes that all reduce NO3 to NO2.
-
Go to “Lists” at the top of the page, title your list, give it a description and a color.
-
Scroll to the bottom, and you’ll see the “List Keywords” menu. Type “Nitrate reductase” and you’ll see a bunch of options pop up; select “nitrate reductase” and then press the “Save list” button.
This list, when used, will search a set of gene annotations for everything containing the words “nitrate reductase”.
Now you’ll see a bunch of information on the next page. On the left-hand side of the page is a box that says “Projects”… next to that, click “Select all”, then on the right of the page click the big blue button that says “Update”. Now you’re looking at all the projects you have access to, and you should see all the nitrate reductase proteins in that set!
Optional: Download sequences for analysis
This isn’t necessary for today’s lab, but will be useful if you want to analyze groups of sequences for your project at the end of the course.
You can download a FASTA file (either DNA or protein) with all of these results by clicking “Download list” (near the top of the page) and selecting the type of FASTA you’d like to download. Feel free to do that with the nitrate reductase results to test it out.
Investigating Metabolic Pathways
Let’s now return to your genome summary.
You can pick any metabolism you are interested in for this lab. As an example, click on “Select lists”, then navigate to “Nitrogen cycle” (if you can’t find it, type it in to the search bar on the left). In the middle bar, right under the words “Nistrogen cycle”, there’s a blue button that says “select all”. Click that, then click “Apply”.
Now you can see the genes predicted to be involved in nitrogen transformation pathways across all your selected genomes! This includes the nitrate reductase genes we were investigating earlier.
Today’s turn-in assignment:
This week each group member should select a genome from your team’s sample and identify at least one pathway or module of interest using ggKbase lists.
What makes a good genome?
- Should be 1.5Mb at least
- Fewer contigs is better
- Ideally the genomes selected by your group should have different taxonomic classifications from one another (but don’t worry too much about this)
You don’t have to each pick a different pathway to study, but you have to pick different genomes of interest for each group member.
Picking a pathway/list
Examples of pathways to consider looking into:
- Methane metabolism
- (Look for mcrA)
- Cellulose degradation
- Aerobic or anaerobic metabolism markers
- Can they utilize oxygen? How do you know?
- Hydrogen utilization
- Look for the hydrogenase large subunit
Once you’ve done this, give me the following information for each genome:
- What is the ggKbase taxonomic classification for this organism?
- How many scaffolds does the genome contain, and what is the total size of the genome bin?
- What pathway (which list) did you investigate?